This page summarizes the chlorine cycle, a portion of which I studied my Ph.D. research using comparative genomics.
Publications:
- The biogeochemical cycling of chlorine. Barnum, T. P., & Coates, J. D. (2022). Geobiology, 00, 1– 16.
- Chlorine redox chemistry is widespread in microbiology. Barnum T.P. & Coates J.D. (2023). ISME J, 17(1), 70-83.
- An uncharacterized clade in the DMSO reductase family of molybdenum oxidoreductases is a new type of chlorate reductase. Barnum, T. P., & Coates, J. D. (2020). Environmental Microbiology Reports, 12(5), 534-539.
- Identification of a parasitic symbiosis between respiratory metabolisms in the biogeochemical chlorine cycle. Barnum TP, Cheng Y, Hill KA, Lucas LN, Carlson HK, Coates JD. ISME J 14, 1194–1206 (2020).
- Genome-resolved metagenomics identifies genetic mobility, metabolic interactions, and unexpected diversity in perchlorate-reducing communities. Barnum TP, Figueroa IA, Carlström CI, Lucas LN, Engelbrektson AL, Coates JD. (2018). ISME J 12: 1568–1581.
Full dissertation:
- Discoveries in the Biology of Oxidized Chlorine. Barnum, TP & Coates, JD (2020). Open-access link.
A biogeochemical cycle for chlorine
There is a common yet mistaken belief that any chemical containing chlorine is unnatural. Chlorine may deserve this reputation: chlorine gas and hypochlorous acid (bleach) are used as disinfectants, perchlorate is a component of explosives, organic chlorine solvents are toxic, organic chlorine plastics are inert, and, most dramatically, chlorine in the atmosphere eats away at the ozone layer protecting all of life on Earth. The chlorine chemicals we synthesize almost seem in direct conflict with nature. Usually the only natural chlorine-containing chemical the average person can name is chloride.
You should understand that, like other elements, chlorine is found in a diverse set of natural organic and inorganic compounds with unique properties, and the distribution of chlorine on Earth is controlled by a biogeochemical cycle (Figure 1).
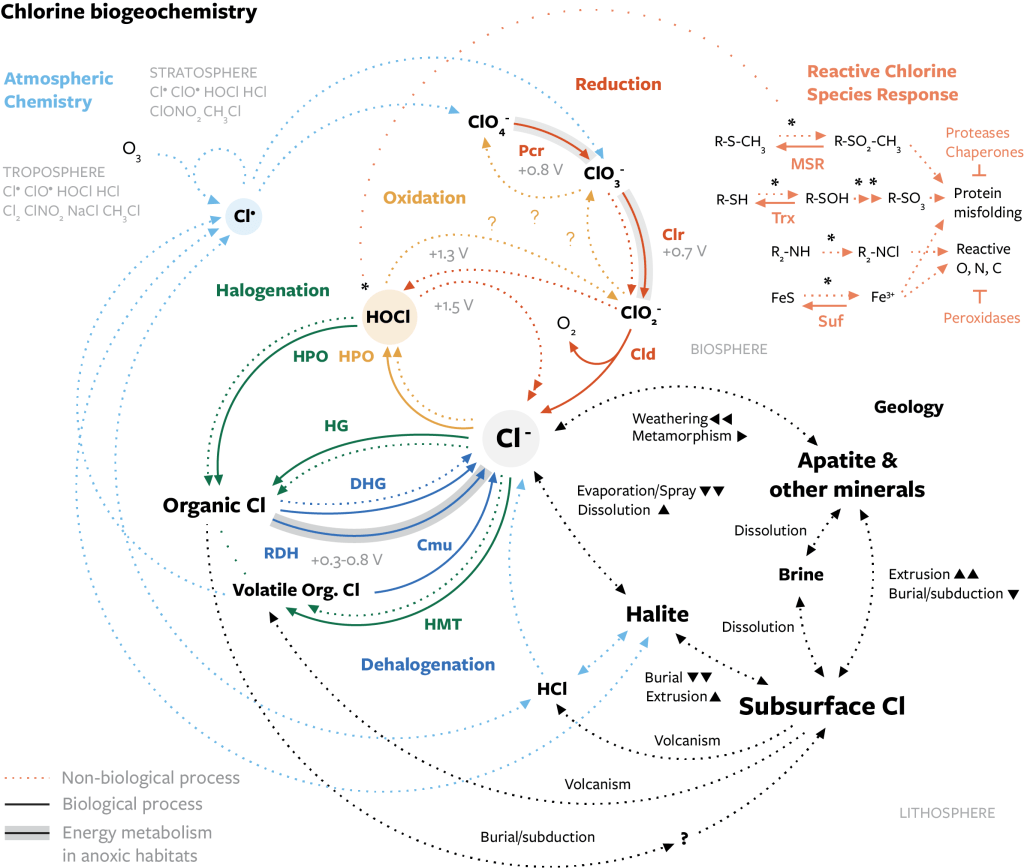
A brief summary of the major processes in the chlorine cycle, which are widespread and occurring continuously:
- Chloride: Chloride is a highly soluble anion that helps solubilize metals in fluids, is subject to strong control by the water cycle, and whose uptake and use as a solute by organisms can control its abundance in systems like soil. The concentration of chloride in the ocean is a major control on its properties and has changed over geologic time due to the sequestration of chloride in evaporite deposits.
- Halogenation: The addition of a chloro group (-Cl) to an organic molecule by enzymes or oxidative chemistry. Various types of life synthesize organochlorine to change the properties of a molecule. Unlike most other elements found in organic molecules, chlorine forms a single bond and more oxidized groups, e.g. a chlorosyl group (-ClO), have not been observed. Organochlorine is often the majority of chlorine in soils and the atmosphere.
- Dehalogenation: The removal of a chloro group, usually by enzymatic activity. Organisms in oxygenated habitats remove chlorine to further catabolize the organic molecule. Organisms in habitats without oxygen reduce organochlorine for a source of energy. Dechlorination can also occur chemically, e.g. through photochemistry.
- Oxidation: Chlorine in water or atmosphere can be oxidized by photochemistry to different oxidation states, including the most oxidized states of chlorine (ClO4– and ClO3–). Oxidation in the atmosphere usually depletes ozone and affects global climate and the ozone layer. Organisms have been observed to oxidized chloride to hypochlorous acid (HOCl) for its potent oxidizing activity. This even occurs within certain cells in the animal immune system.
- Reduction: Oxidized chlorine compounds have high reduction potentials, and their reduction is often spontaneous. Microorganisms in low-oxygen habitats reduce oxidized chlorine for a source of energy.
- and more: cellular responses to oxidized chlorine compounds, interactions with other halogen elements, etc.
The myriad biological, chemical, and geological processes in chlorine cycle have been largely – and in my view wrongly – discussed separately. In the course of my own research, I learned that I needed to consider the interconnectedness of oxidized chlorine compounds with other processes in the chlorine cycle, which resulted in this review.
I challenge scientists to understand how chlorine behaves in nature and may be involved in their study system.
Oxidized chlorine: perchlorate and chlorate
Perchlorate (ClO4–) and chlorate (ClO3–) form naturally in the atmosphere. Anthropogenic sources of these compounds make them concerning pollutants. When oxygen is low, certain bacteria reduce these compounds to provide energy using specialized metabolic pathways. Many of these organisms have been isolated and studied in the laboratory to discover what genes are required for the pathway and how those genes evolved.
What do we know about perchlorate- and chlorate-reducing bacteria?
Prior to my work, science had described the biochemistry, genetics, physiology, and application of bacteria performing these metabolisms. Perchlorate reductase is specialized enzymes for perchlorate reduction, whereas chlorate reductases appeared more promiscuous. What made a chlorate reductase a chlorate reductase was the expression of a chlorite dismutase that degrades the produced chlorite (ClO2–) into chloride and molecular oxygen. Genes for these enzymes can be acquired through horizontal gene transfer and are usually expressed when better respiratory electron acceptors, like oxygen or nitrate, are unavailable. It also appeared that under certain circumstances the pathway could be split between different cells, with some cells reducing pechlorate and chlorate to chlorite, and other cells degrading the chlorite.
What do communities look like when perchlorate is the main source of energy?
Because many bacteria cannot be grown in the laboratory, it was suspected that wild communities degrading perchlorate had more diverse genes, organisms, and interactions than had been previously found.
In 2018 [1], I studied communities of microorganisms from the San Francisco Bay that degraded perchlorate by assembling genomes from metagenomes (which involves sorting a mixture of sequences from different organisms into distinct genomes). I also used assembly graphs to link horizontally transferred genes to specific genomes, which was a new addition to the field that has since been improved with automated methods. As expected, communities reducing perchlorate had a greater diversity of organisms than identified before. Based on the metabolisms of all genomes present, there are likely interactions between chlorine and sulfur-cycling organisms that influence rates of perchlorate reduction. Another type of interaction observed was the presence of small bacteria that attached to another bacterium (see later discussion here).
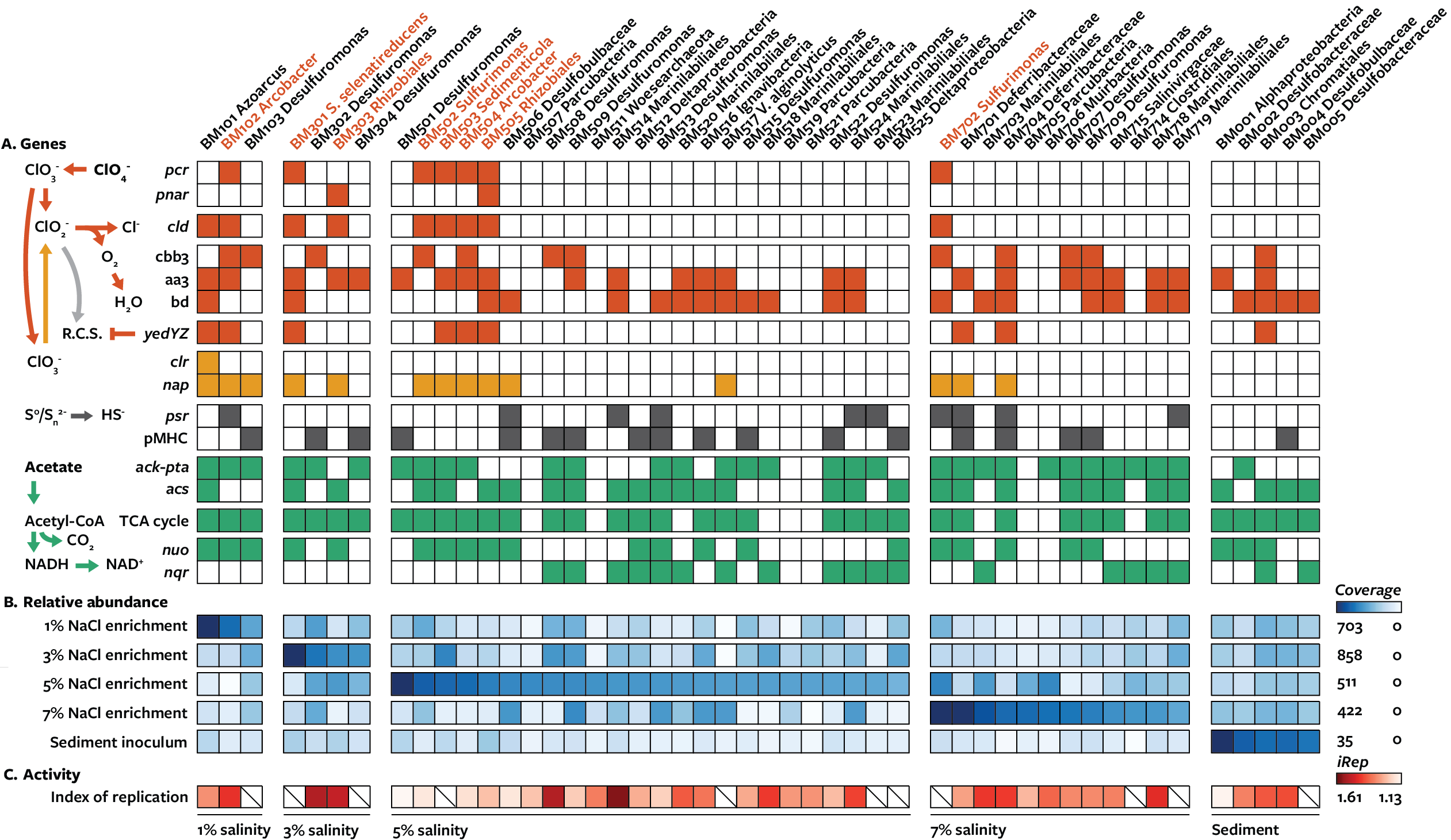
Some organisms had a novel enzyme that comparative genomics, protein alignments, and phylogenetics suggested involvement in this process, which I later demonstrated to be the third type of chlorate reductase using comparative genomics and lab assays. (2nd publication below)
What are specific metabolic interactions that occur during perchlorate and chlorate reduction?
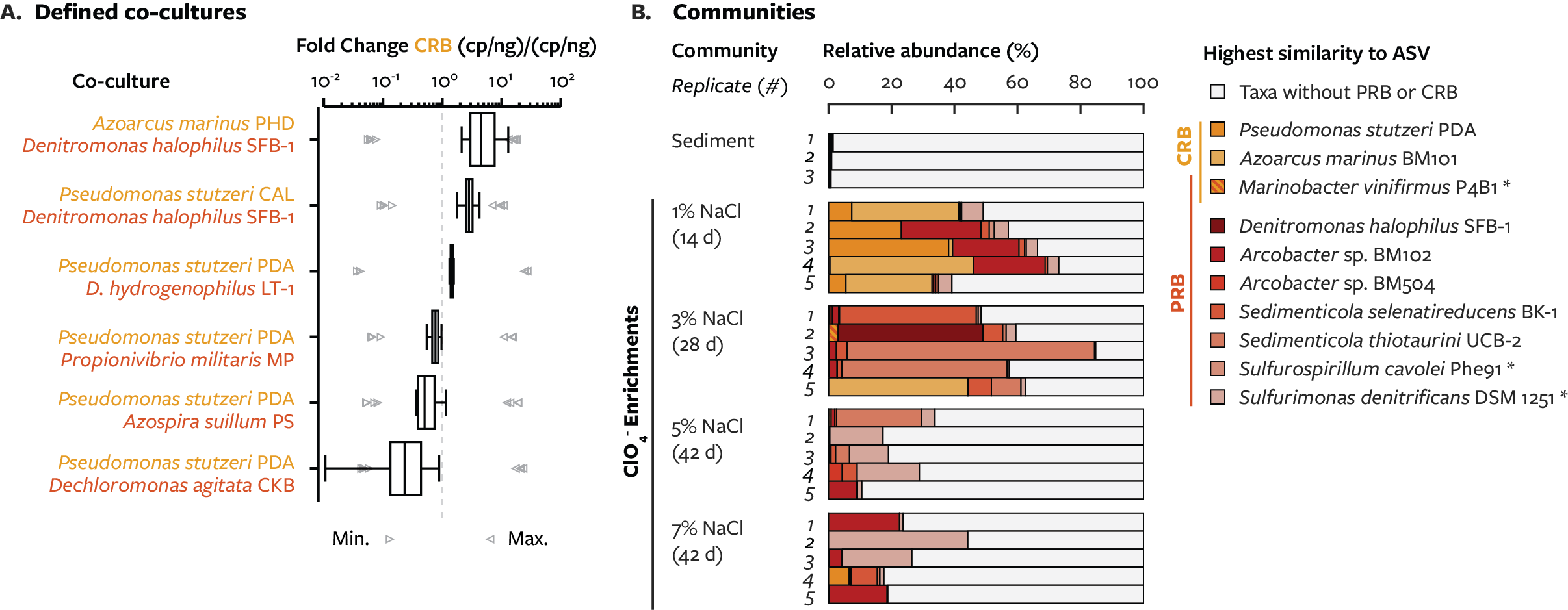
A chlorate-reducing bacterium had been identified in perchlorate-reducing communities [1], raising the intriguing possibility that these metabolisms interact but without direct confirmation. Chlorate (ClO3–), chlorite (ClO2–), and oxygen (O2) are the intermediates in the biochemical pathway for reducing perchlorate to chloride and water. Each of those compounds – but not perchlorate – can be used by chlorate-reducing bacteria.
While isolating perchlorate-reducing bacteria, a process that should result in one single organism in culture, I discovered that cultures were regularly contaminated by chlorate-reducing bacteria that were altering the rate of perchlorate reduction. Beyond simply persisting in the culture, genomic sequencing showed that these chlorate-reducing bacteria dominated their small community. To understand the mechanism of the interaction, I used genetic deletion of pathway genes to measure phenotypes in co-cultures and explain the co-culture growth kinetics. This discovery greatly improves of our understanding of how perchlorate and chlorate are degraded in nature.
Data availability
- Code: github.com/tylerbarnum
- Genomes and sequencing reads: NCBI BioProject PRJNA387015
Oxidized chlorine: chlorite and hypochlorous acid
The lower oxidation states of chlorine are more reactive than the higher oxidation states. Chlorite (ClO2–) and hypochlorous acid (HOCl) are strong oxidants that react with various biological molecules. Chlorite is not known to be produced naturally from oxidation, but one place where it is an important consideration is as a residual from the use of chlorine dioxide (ClO2) in water treatment. Hypochlorous acid is understood to have important biological roles in many environments. For example, our immune system oxidizes chloride to hypochlorous acid for its efficiency in killing bacterial pathogens, and organisms in soil oxidize chloride to hypochlorous acid to degrade organic matter.
Evidence for the broad presence of chlorite from genomes and metagneomes
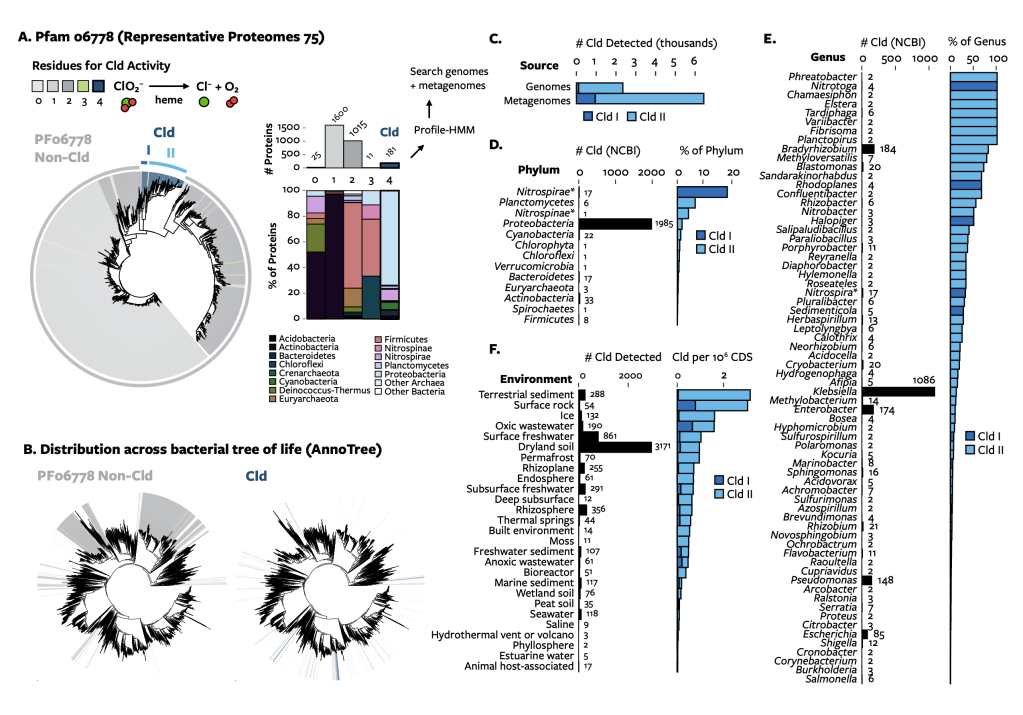
A chlorite-degrading enzyme called chlorite dismutase (Cld) is one of several bacterial genes known to respond to reactive chlorine stress. It is only used to degrade chlorite. Unlike hypochlorous acid, chlorite is not known to be common, and the prevalence of this specific enzyme in hundreds of bacteria has left scientists perplexed.
I approached the problem from a comparative genomics perspective. A search identified Cld in far more genomes and metagenomes than are expected based on the enzyme’s only known role, in perchlorate and chlorate reduction. Various taxa had Cld, and horizontal gene transfer across geologic time to the present was apparent from its phylogeny. Cld was linked with numerous genes that have plausible roles in the biology of oxidized chlorine: perchlorate and chlorate reductases, oxidative stress repair, transporters, signaling proteins, and more. The result is the most comprehensive analysis of genes involved in reactive chlorine stress yet identified. The comparative genomics of Cld also suggested sources of chlorite include co-metabolic reduction (i.e. perchlorate reduction by a nitrate reduction pathway) and possibly the oxidation of chlorine, which has not been seen before naturally.
Outlook
The most exciting new discoveries in microbiology will involve both experimentation and mining large sets of genomes and metagenomes. Using this approach, my research has identified new enzymes and interactions that participate in converting oxidized chlorine to chloride, and an unexpectedly large influence of oxidized chlorine species in nature. These findings provide a clearer picture of how microorganisms participate in the biogeochemical chlorine cycle and will support research in fundamental and applied environmental microbiology. Research that defines the mechanisms of chlorine oxidation and further understands responses to reactive chlorine species would be more helpful for understanding the biological importance of chlorine amid the other elements the life encounters.